Benchmark#
The torchtree software was evaluated alongside other phylogenetic tools in a published benchmark study [1].
This benchmark assesses the memory usage and speed of various gradient implementations for phylogenetic models, including tree likelihood and coalescent models. The study’s aim was to compare the efficiency of automatic differentiation (AD) and analytic gradient methods. The gradient of the tree likelihood can be computed by BITO or physher [2], efficient C++ and C libraries that analytically calculate the gradient. torchtree integrates with these libraries through the torchtree-bito and torchtree-physher plug-ins. physher is a standalone program that can be used as a library by torchtree.
Program |
Language |
Framework |
Gradient |
Libraries |
|---|---|---|---|---|
C |
analytic |
|||
Stan |
Stan |
AD |
||
python |
JAX |
AD |
||
python |
PyTorch |
AD |
BITO and physher |
|
python |
TensorFlow |
AD |
In this study, we compared six gradient implementations of the phylogenetic likelihood functions, in isolation and also as part of a variational inference procedure. The data consisted of a collection of influenza A datasets ranging from 20 to 2000 sequences sampled from 2011 to 2013.
This macrobenchmark simulates the core steps of a real phylogenetic inference algorithm but simplifies the model to make it easier to implement across different frameworks. In this setup, we are estimating parameters of time tree under a strict clock with a constant-size coalescent model. Each implementation relies on automatic differentiation variational inference (ADVI) to maximize the evidence lower bound (ELBO) over 5,000 iterations. We specify an exponential prior (mean = 0.001) on the substitution rate and the Jeffrey’s prior for the unknown population size.
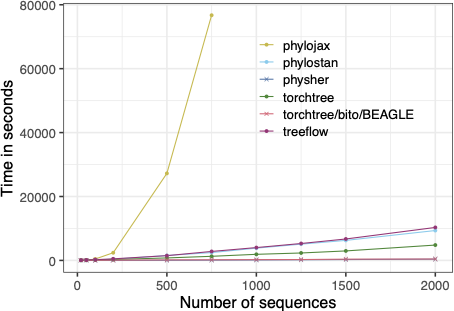
Speed of implementations for 5,000 iterations of variational time-tree inference with a strict clock.#
As shown in the next figure, the relative performance of AD depends on the task.
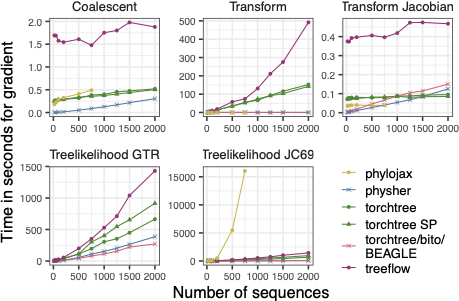
Speed of implementations for the gradient of various tasks needed for inference.#